Cloning for all #2 - Golden Gate
Published:
On the second installment of #CloningForAll we look at Golden Gate, or seamless restriction cloning with nearly unlimited fragments! 1/n
What is Golden Gate?
Golden Gate is restriction cloning with fancy restriction enzymes.
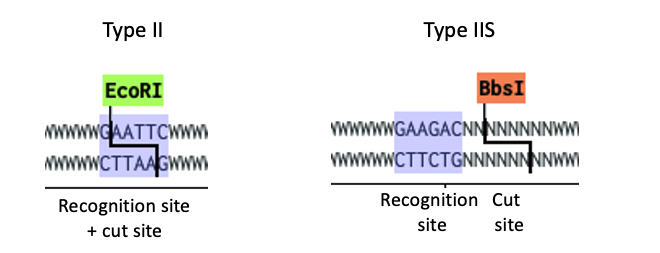
- The restriction enzymes we all know and love e.g. EcoRI are Type IIP (for palindromic since their recognition site is a palindrome) cut where they bind, every time you cut with EcoRI you create the same overhangs so you can only put together two fragments cut with EcoRI. Every time you ligate two pieces cut with EcoRI you reconstitute the EcoRI site between them.
- Golden Gate is done with Type IIS (Shifted cleavage) enzymes that recognize one site, and cut away from it. They don’t care about the sequence of their cut site, so we can use them to create any overhang we need. Instead of having the recognition site between the two fragments we have seamless assemblies!
Why Golden Gate?
Like Gibson assembly, Golden Gate opens up our world to multi fragment (30++) seamless assemblies. If you are trying to introduce multiple fragments with homology Golden Gate is much liklier to work because it uses simple ligation of overhangs. I (and others) have found that the Gibson chewback can expose similar regions and create chaos in these situations. Finally, the enzymes required for Golden Gate are cheaper at scale, so if you have a large cloning project (especially if some fragments are common between plasmids) it is a much more economical option.
Unlocking the potential of Type IIS Restriction Enzymes
The true magic of Golden Gate is that you cut away from the recognition site. I am going to use an example that uses Type IIS enzymes to show how they work and then we will get into true Golden Gate. CRISPR, it won a nobel, everybody does it these days. The machinery to express sgRNAs is usually a U6 (250 bp) + guide (20 bp) + scaffold (80 bp) + 6Ts to stop DNA Pol III. Since only 20 bp of that ~350 bp sequence need to change, there’s no reason to put that all together every time. Enter Type IIS. Between the U6 and scaffold are two outward facing BsaI sites. i.e. the enzyme sits on the two highlighted portions and cuts outwards towards the U6 and scaffold.
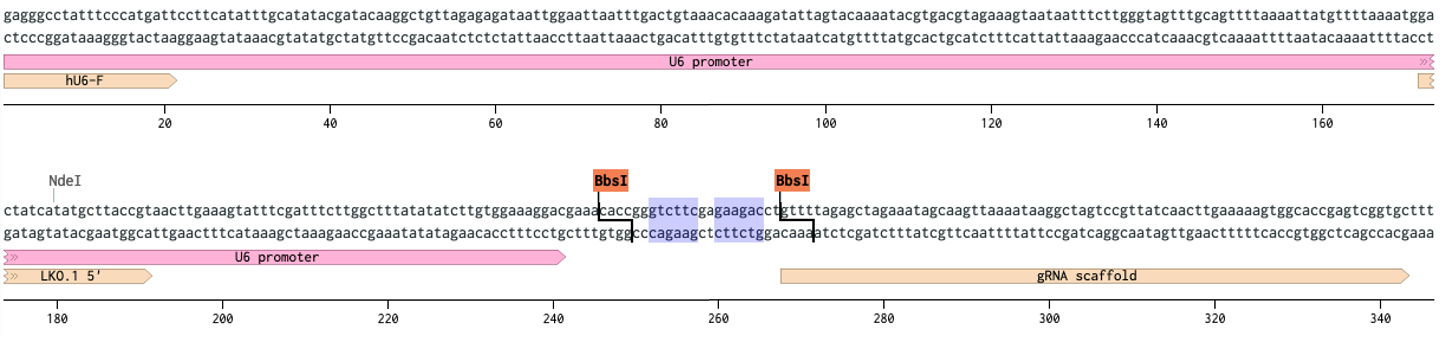
When you cut the 2x BsaI fragment drops out leaving unique overhangs. We introduce our guide sequence by annealing two oligos that have the correct overhangs, seamless assembly so that your guide starts at the right base. The position of the guide sequence with regards to the U6 and scaffold are important for its expression and function, so you can’t afford to use Type IIP restriction sites that either 1) limit your options sequence wise, and 2) get in the way.
For a full description of the protocol you can find a great write up by Addgene referencing the Zhang lab protocol.
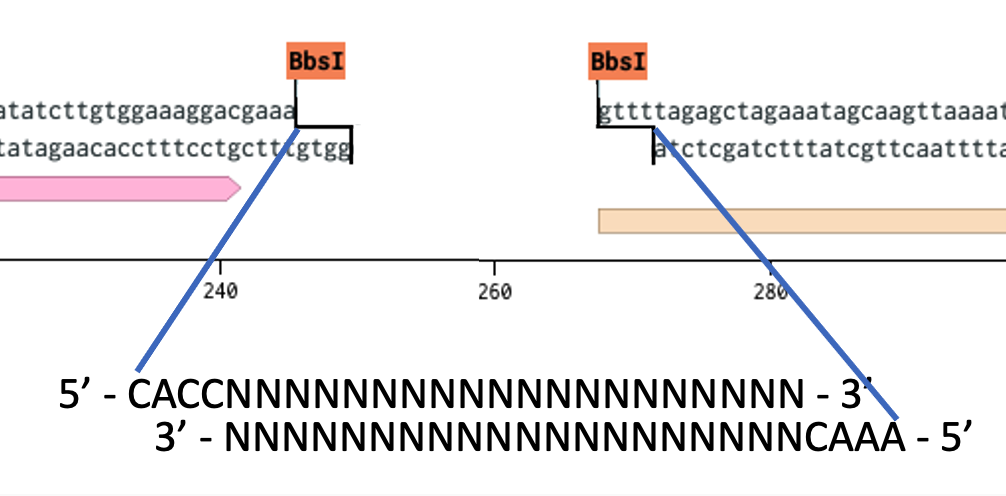
It’s worth mentioning here that while this approach for guide design has served many well it requires a lot of steps and reagents. Annealing, phosphorylation, etc. I have become a fan of single-stranded Oligo Gibson assembly with NEB HiFi for these kinds of cloning projects. A single oligo and you are good to go. This works great for single guide drops, but where it truly shines is for plasmid library preparations. i.e. when you are trying to introduce a large amount of different sequences into the same drop site to create a library of plasmids. For the full protocol see NEB’s website
Let’s look at Golden Gate
Similar to the guide dropzone, Golden Gate uses recognition sites that create a unique overhang and also cut the recognition site out of the fragment. By creating overhangs that are unique to each joining site you can bring the fragments together in the correct order.
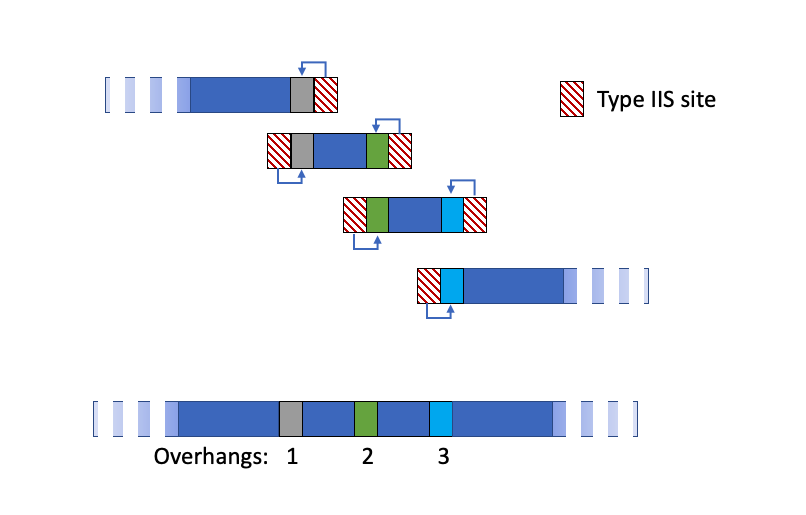
The protocol of Golden Gate is repeated rounds of cutting at 37C, ligating at 16C, and ends on a cut at 37C! When the correct fragments ligate, they don’t reintroduce a cut site and don’t cut in the next cycle. But if a restriction end ligates back on, it will be cut in the next cycle and give the fragments another chance to come together correctly during the ligation. The final cutting ensures that anything that isn’t what you want is linear and won’t be propagated.
Planning your cloning
You’ve seen most of my tricks in the Gibson assembly thread I will try to give you a visual of how to create the matching overhangs between fragments so you can understand how this all works. The first step is to make sure that the backbone and fragments dont have the Type IIS enzyme you plan to use (and hopefully avoid spending a significant amount of time finding all the sequences just to find that all the possible Type IIS sites are unusable). This is admittedly one of the drawbacks of Golden Gate and a reason Gibson is easier to design. Recently NEB came out with a new enzyme to try to overcome this PaqCI has a 7 bp recognition site (instead of 6) and is therefore rarer.

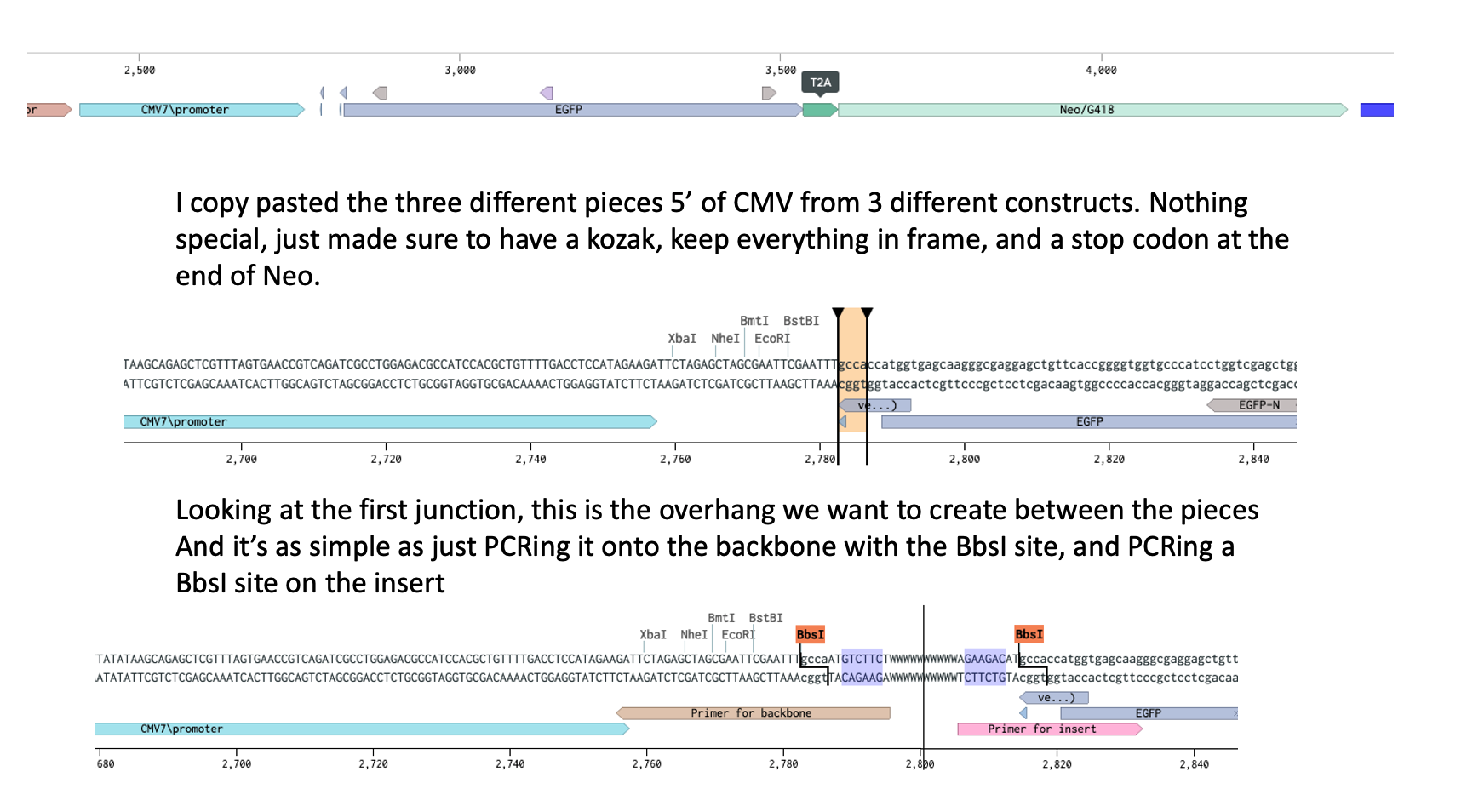
You’ve seen how to add the same overhang to both ends, one of them should already have it, the other one just needs it PCRed on. Now you do it for all the fragments. I’m a control freak and like to be able to know exactly what happens to my cloning so I do it manually. Normal humans use NEBridge
NEB has some great resources for this, especially for which enzymes/buffers can be mixed. I also found this protocol really helpful when first starting out http://parts.igem.org/wiki/images/e/eb/Rice_igem_protocols.pdf
Wrapping it all up
Gibson and Golden Gate head to head. Both are great for complex, multi-fragment, seamless constructions. Golden Gate handles repetitive sequences in fragments much better (an example of this is trying to put 3 sgRNA cassettes in with U6+scaffold, good luck doing that with Gibson). Golden Gate is arguably cheaper since it just uses a little restriction enzyme + ligase.
Gibson has strengths, it’s arguably easier to design. For both these approaches we looked at PCRing the whole backbone to introduce homology arms (Gibson) or correct overhangs (Golden Gate). But with Gibson you don’t have to PCR the backbone, you can cut the backbone with 1 or 2 restriction sites that are generally where you need them and PCR homology for the backbone onto the fragments. With the NEB HiFi chewback you can also remove or modify the drop in sites to make it seamless.
And finally, for single drop ins, and especially libraries. Single stranded oligo Gibson is 100% the way to go.
Fun literature: 35 fragment assembly with golden gate
And that’s a wrap. Hope you found something useful here. Let me know if there’s anything you’d like me to cover! Thanks for reading.
